The immune response to infection by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the cause of the ongoing coronavirus disease 2019 (COVID-19), consists of both humoral and cellular responses. Serum antibodies against the virus neutralize it by preventing its entry into the cell and the successful establishment of infection. Other antibodies help to clear the virus from the body.
A new preprint research paper posted to the bioRxiv* server provides a greater understanding of the aspect of humoral immunity against the virus – the plasma cells that pour out a flood of specific antibodies targeting various epitopes on the pathogen.
Spike antigen
The major viral antigen is the spike glycoprotein, composed of the S1 and S2 subunits. The S1 subunit contains the receptor-binding domain (RBD) that mediates viral attachment to the angiotensin-converting enzyme 2 (ACE2) receptor on the host cell. The S2 subunit is responsible for viral fusion to the cell membrane, following further processing by the host protease TMPRSS2 and internalizing the virus.
S1 is the target for many immunoglobulin antibodies, which are often neutralizing since antibodies to the RBD are likely to interact with the virus's interface and the ACE2 receptor, inhibiting viral entry into the cell. TMPRSS2 inhibitors may also disrupt the priming of the virus, thus preventing its entry.
Plasma cells in SARS-CoV-2 infection
Convalescent plasma containing high titers of neutralizing antibodies and several therapeutic monoclonal antibodies directed against the RBD, and isolated from the B cells of COVID-19 patients, are being used to treat COVID-19. These include bamlanivimab and etesevimab (Eli Lilly), with many others being in advanced clinical trials.
It is relatively easy to isolate antibodies specific to the virus by screening memory B cells because they have B cell receptors (BCR) on their surface. Soluble forms of the specific antigens to which these BCRs are cognate, such as the spike or RBD, can be used to label the required subset of antigen-binding cells and sort them by flow cytometry.
However, antigen-specific B cell clones make up less than one in a thousand of the total memory B cells in COVID-19 patients, while the need to sort and clone single cells further reduces the yield.
Importantly, plasma cells (PCs) produce antibodies detected in serum rather than memory B cells. Recent research has shown that many antibodies closely related to SARS-CoV-2 show low somatic hypermutation rates, similar to the germline antibodies.
Thus, the early antigen-specific plasma cells in peripheral blood are possibly encoding specific and neutralizing antibodies to the virus, despite the apparent lack of maturation.
The transitional plasma cells are of different types, from plasmablasts to short-lived PCs, found in blood only briefly before migrating into the bone marrow to form long-lived PCs. While transitional PCs make up only 5% of B cells in the peripheral blood, they increase to almost a fifth during COVID-19. However, memory B cells make up 40-50% of total B cells both before and after infection with SARS-CoV-2.
Clonally expanded PCs in people exposed to an antigen have been associated with specific binding to target antigens, but the relevance of this finding to COVID-19 patients remains to be verified.
Study details
The current study began with the development of a protocol to select SARS-CoV-2 spike antigen-specific plasma cells. This was necessary because of the inability to select PCs that produce specific antibodies using the available enrichment methods since these depend on the expression of BCRs for various immunoglobulins on the cell surface.
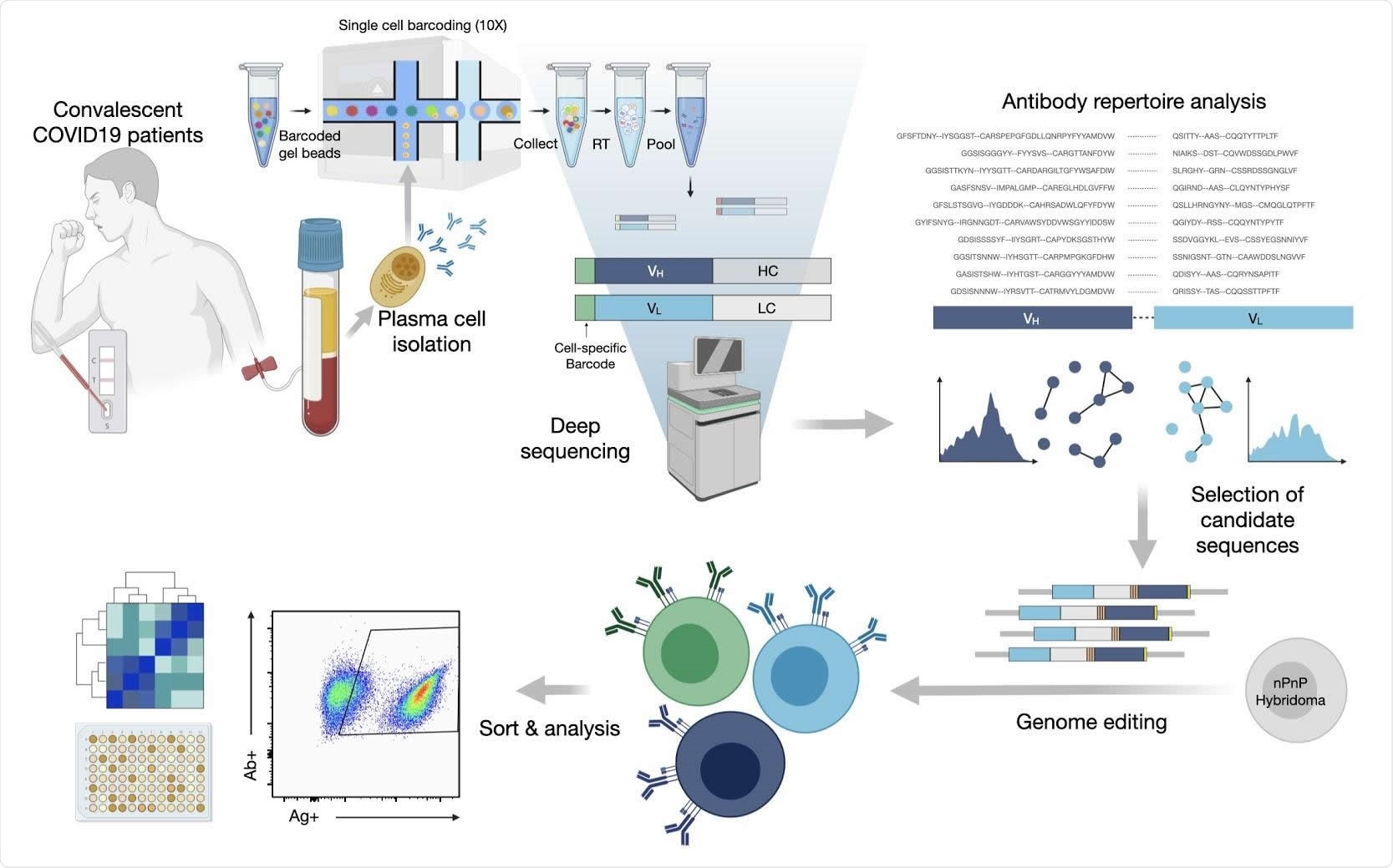
The researchers first used their protocol to sequence the repertoire of antibodies in single PCs. They were able to identify the expanded clones of PC lineages using variable heavy and variable light chain sequences (VH and VL, respectively). The total antibody repertoires, as well as the expanded clones, displayed the use of a broad range of germline genes and sequences.
They identified 45 antibodies with 80% similarity of amino acid sequence in the complementarity determining region (CDR) H3, forming a clonal lineage.
They rapidly discovered that synthesizing thousands of synthetic antibody genes corresponding to the expanded antibody sequences was not feasible, limiting their use of this tool. They nevertheless set out to answer the question as to whether PCs showing high clonal expansion produced specific and neutralizing antibodies in COVID-19.
They selected 132 expanded PC clones with the most abundant clonal lineages. These showed broad sequence diversity. As expected, the immunoglobulin class-switched antibody sequences had over 94% identity with germline genes, on average, with 26 of them showing 100% identity.
These were inserted into a mammalian cell antibody surface display screening system by genome editing, using the CRISPR-Cas9 platform. In this way, they introduced synthetic antibody genes into the endogenous IGHV locus on the genome.
Specific antibodies
They found 37 unique antibodies that were specifically directed against the SARS-CoV-2 antigens, arising from unique expanded clones from 11 patients. Of these, 11 were identical to the germline. Thus, most of the COVID-19 patients had PCs from highly expanded clones, producing antibodies specific to the virus. Two patients had no anti-S antibodies at all.
The investigators found that two of the monoclonal antibodies identified here had been reported in earlier studies.
All patients did not show the same degree of specificity to the S1 or S2 subunits due to undetectable antibody levels against the S protein. Also, clonal expansion could not be identified in some cases because of the low single-cell sequencing depth.
The two antibody libraries created by the display platform contained cross-reactive antibodies that bound other human seasonal coronaviruses, but not the highly pathogenic SARS-CoV or MERS (Middle East Respiratory Syndrome)-CoV. This is in contrast to the broader cross-reactivity found in other studies.
The specific antibodies to the SARS-CoV-2 antigens failed to show a familiar repertoire or uniform sequences, indicating a broad spectrum of antibody repertoires in this infection. However, the researchers did find similarity of sequences to those reported by earlier researchers. This indicates that the germline genes drive antibody responses to SARS-CoV-2 infection.
Again, only anti-S antibodies were screened, excluding those that bind to the nucleocapsid protein. Again, the change in the antibody construct format may have affected antibody affinity, stability and expression, causing a loss of reactive sequences.
The researchers comment, “The overall number of antibodies that were identified to be specific for SARS-CoV-2 in this study is likely an underestimation, as not all of the pooled candidates were tested separately.”
Neutralizing antibodies
Three of these antibodies were found to have potent neutralizing activity against the RBD of SARS-CoV-2, using a pseudoviral assay. Two of them have previously been described in other studies. Again, two of the three were 100% identical to germline antibodies but were specific to the RBD.
Two of these appeared to bind overlapping epitopes, and the third a distinct epitope.
What are the implications?
“Our integrated workflow of single-cell sequencing and mammalian display screening is able to demonstrate that convalescent COVID-19 patients produce highly expanded PCs with specific and neutralizing antibodies for SARS-CoV-2.”
Though limited, these findings serve as a proof of concept, indicating this pipeline's ability to identify specific antibodies with high binding affinity and potent neutralization capacity, using a single-cell sequencing approach on peripheral blood PCs.
In a pandemic, or when an infection requires to be diagnosed early when the antigen is not available for testing, or in the presence of a low number of memory B cells against specific antigens, such an approach could be very beneficial. PCs in peripheral blood are also the only source of serum antibodies, from which therapeutic antibodies are derived.
With refinements in the current selection process for clonally expanded lineages, a more significant number of antigen-specific antibodies could possibly be identified using this technology.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Ehling, R. A. et al. (2021). Single-cell sequencing of plasma cells from COVID-19 patients reveals highly expanded clonal lineages produce specific and neutralizing antibodies to SARS-CoV-2. bioRxiv preprint. doi: https://doi.org/10.1101/2021.02.12.430940. https://www.biorxiv.org/content/10.1101/2021.02.12.430940v1
Posted in: Medical Research News | Disease/Infection News
Tags: ACE2, Amino Acid, Angiotensin, Angiotensin-Converting Enzyme 2, Antibodies, Antibody, Antigen, Assay, binding affinity, Blood, Bone, Bone Marrow, Cas9, Cell, Cell Membrane, Cell Sorting, Cloning, Convalescent Plasma, Coronavirus, Coronavirus Disease COVID-19, CRISPR, Cytometry, Enzyme, Flow Cytometry, Genes, Genome, Genome Editing, Germline, Glycoprotein, High-throughput screening, Immune Response, Immunoglobulin, Locus, Pandemic, Pathogen, Protein, Receptor, Research, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Virus

Written by
Dr. Liji Thomas
Dr. Liji Thomas is an OB-GYN, who graduated from the Government Medical College, University of Calicut, Kerala, in 2001. Liji practiced as a full-time consultant in obstetrics/gynecology in a private hospital for a few years following her graduation. She has counseled hundreds of patients facing issues from pregnancy-related problems and infertility, and has been in charge of over 2,000 deliveries, striving always to achieve a normal delivery rather than operative.
Source: Read Full Article