A team of Brazilian scientists has recently conducted genomic and phylogenetic analyses of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genome to understand the viral evolution process better. Their analyses reveal that the E484K mutated variant of SARS-CoV-2 is present in three different lineages in four Brazilian regions and that the mutation may serve as a common solution for viral evolution. The study is currently available on the bioRxiv* preprint server.
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative pathogen of coronavirus disease 2019 (COVID-19), is a member of the Coronaviridae family, which are enveloped positive-sense, single-stranded RNA viruses with a larger genome size of about 30 kb. Since its emergence in December 2019 in China, researchers worldwide have sequenced almost 4,10,000 genomes of SARS-CoV-2 and have shared the information on the Global Initiative on Sharing All Influenza Data (GISAID) database.
Several genomic epidemiological studies of SARS-CoV-2 have been conducted, with the majority focusing on the mutations in viral spike protein, a surface glycoprotein of SARS-CoV-2 responsible for binding with the host angiotensin-converting enzyme 2 (ACE2) and initiating the viral entry process. Studies are currently mainly focusing on three viral lineages, B.1.1.7, B.1.351, and P.1. because of their significant impact on viral transmissibility and pathogenicity. Of these lineages, lineage P.1 is prevalent in Brazil with three mutations (K417T, E484K, and N501Y) in the spike receptor-binding domain (RBD). In Brazil, many viral genomes harbor E484K mutation, which is associated with increased viral transmissibility and immune evasion.
In the current study, the scientists evaluated the mutation characteristics and phylogenetic relationships in currently known lineages in Brazil to understand the adaptive evolution process of SARS-CoV-2.
Important observations
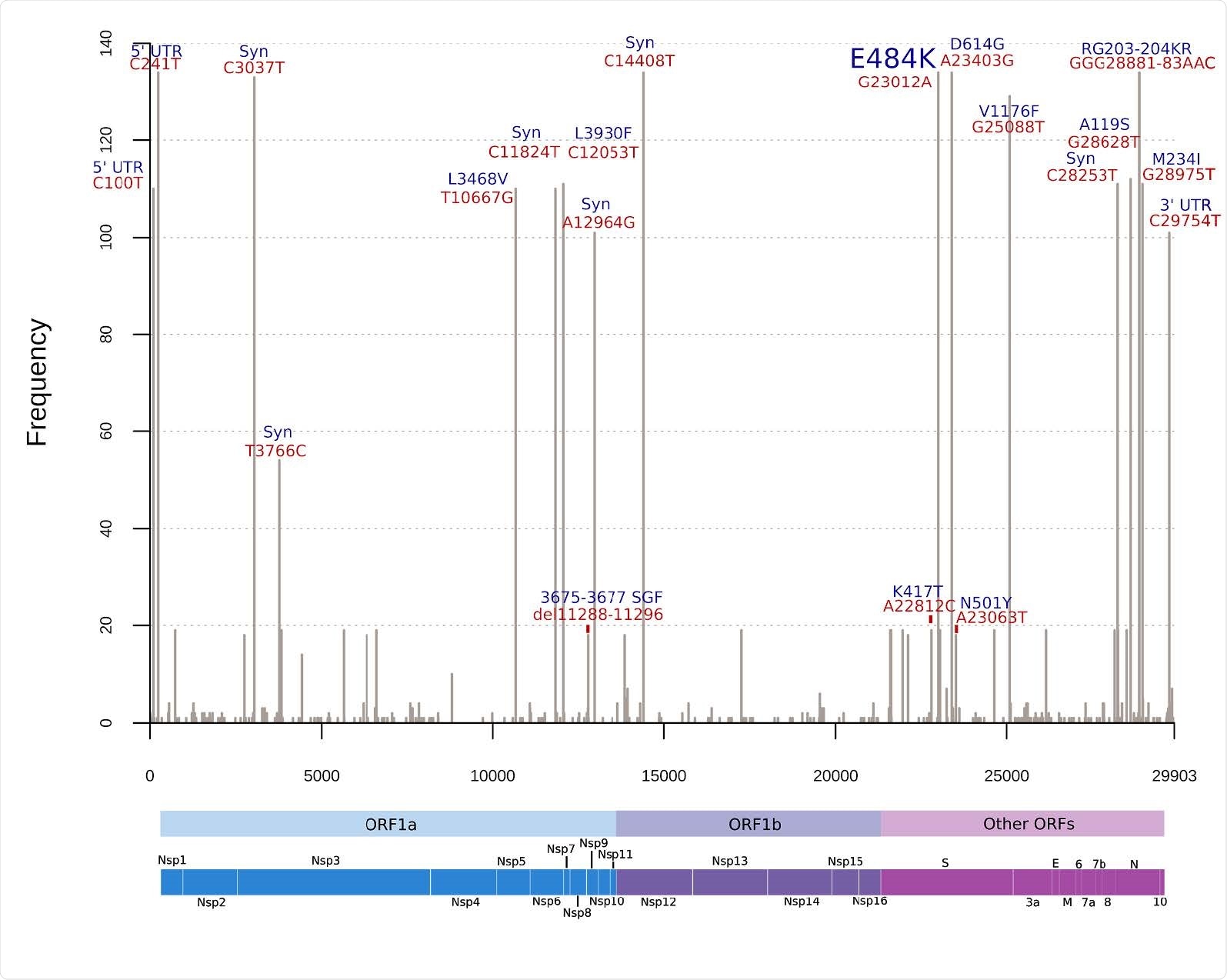
The study findings revealed that SARS-CoV-2 variants with E484K mutation are widely distributed in many regions in Brazil. The mutation was introduced in Brazil in October 2020. Because the E484K mutation is found in different viral lineages simultaneously with other mutations, the scientists suggest that this particular amino acid substitution may act as a common driving force for viral evolution in different genetic variants of SARS-CoV-2. In the E484K mutation, a negatively charged amino acid (glutamic acid) is substituted with a positively charged amino acid (lysine). Thus, it can be expected that the mutation has a significant impact on viral sustainability and adaptive evolution.
The structural analysis revealed that a new site for ACE2 binding (amino acid 75) is generated because of the E484K mutation. This appears to create a significantly stronger interaction between ACE2 and the native binding site located at the RBD and ACE2 interface (amino acid 501).
A highly diverse range of genetic mutations was observed in all Brazilian lineages with E484K mutation. On average, about 19 and 30 mutations were observed in B.1.1.33 and P.1 lineages, respectively. Further genomic analysis of the most recently emerged lineage P.2 indicated that both P.1 and P.2 lineages are evolving rapidly and have been circulating in Brazil for a more extended period.
The analyses of new specific single substitutions were also carried out in the current study. One such mutation is A27V, present in the N-terminal domain (NTD) of the spike protein. Studies have suggested that mutations in the spike NTD can allosterically alter the spike RBD – ACE2 interaction dynamics and facilitate immune evasion. Moreover, evidence shows that viral lineages carrying E484K mutation are associated with increased escape from antibody-mediated neutralization. Although the actual impact of E484K and other substitutions on immune evasion is still not clear, the scientists suggest that E484K mutation is associated with increased viral fitness under natural selection pressure.
Study significance
Overall, the study indicates that the E484K mutation acts as a critical evolutionary event for different viral lineages in terms of increasing viral fitness. The E484K mutation could potentially increase the infectivity and immune evasion potency of SARS-CoV-2. According to the scientists, more studies investigating the effectiveness of anti-SARS-CoV-2 human sera against E484K mutated variants of SARS-CoV-2 are urgently needed.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- bioRxiv preprint server. 2021. Ferrareze PAG. E484K as an innovative phylogenetic event for viral evolution: Genomic analysis of the E484K spike mutation in SARS-CoV-2 lineages from Brazil. https://www.biorxiv.org/content/10.1101/2021.01.27.426895v1
Posted in: Medical Research News | Disease/Infection News
Tags: ACE2, Amino Acid, Angiotensin, Angiotensin-Converting Enzyme 2, Antibody, Coronavirus, Coronavirus Disease COVID-19, Enzyme, Evolution, Genetic, Genome, Genomic, Glutamic Acid, Glycoprotein, Influenza, Lysine, Mutation, Nucleotide, Pathogen, Protein, Receptor, Respiratory, RNA, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spike Protein, Structural Protein, Syndrome

Written by
Dr. Sanchari Sinha Dutta
Dr. Sanchari Sinha Dutta is a science communicator who believes in spreading the power of science in every corner of the world. She has a Bachelor of Science (B.Sc.) degree and a Master's of Science (M.Sc.) in biology and human physiology. Following her Master's degree, Sanchari went on to study a Ph.D. in human physiology. She has authored more than 10 original research articles, all of which have been published in world renowned international journals.
Source: Read Full Article