Can the specificity of DNA be harnessed?
DNA has very high specificity, so that a particular DNA sequence will always bind to its complementary DNA sequence. This can, therefore, be used to design nanomachines with high specificity by using specific DNA sequences. It is even possible to create branches off a DNA double helix, which makes it possible to create 3D structures.
It can be considered that some biological processes are driven by biological machines, and researchers have sought to use naturally occurring machines as the basis of molecular machines that can potentially connect the molecular world to the macroscopic world.
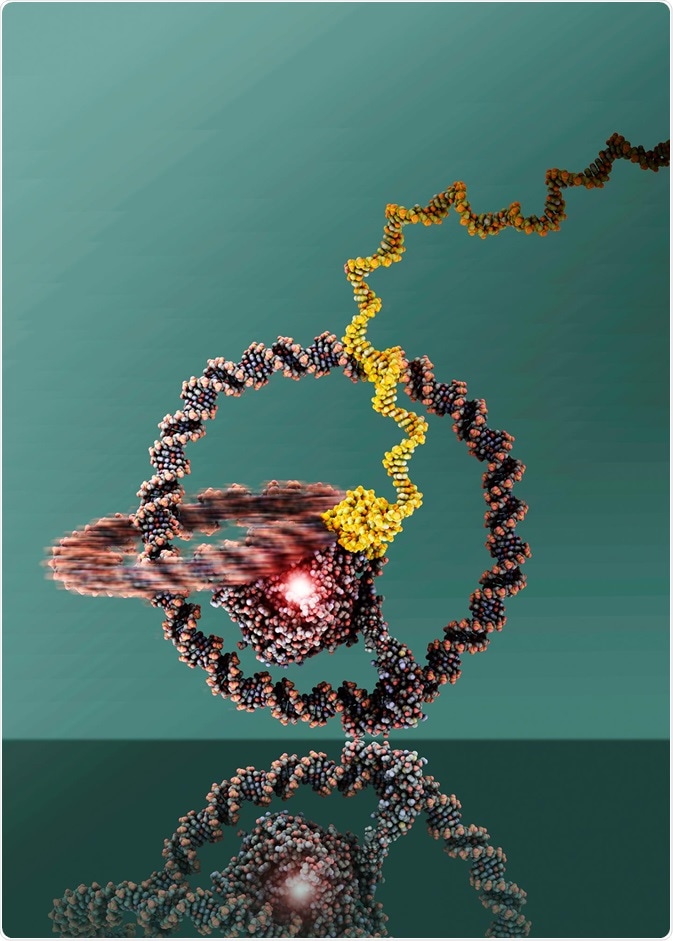 In 2019, researchers built a one-wheeled vehicle out of DNA rings. The two rings are linked like a chain and can well be recognized. At the center is T7 RNA Polymerase. Image Credit: Julian Valero, The University of Bonn.
In 2019, researchers built a one-wheeled vehicle out of DNA rings. The two rings are linked like a chain and can well be recognized. At the center is T7 RNA Polymerase. Image Credit: Julian Valero, The University of Bonn.
DNA nanomachines?
Because it is possible to precisely shape DNA, it is feasible to position a functional group in a precise location; for example, this has been done to create molecular electronic circuits, near-field optical devices and enzyme networks. So, is it possible to make biological nanomachines based around DNA?
DNA is not the obvious choice for biological nanomachines. Larger structures like proteins are able to carry out catalytic reactions and RNA has the ability to form weak bonds and thus secondary structures, which in turn can stabilize structures. However, DNA has an advantage that its structure is simple, therefore it is easy to control how the nanomachine is assembled.
Types of DNA nanomachines
Molecular switches
This is the simplest of the DNA nanomachines, which alternate between two states according to changes in its environment, or by signaling.
Rotational movement can be produced from DNA by changing how the DNA strand twists. C-G nucleotides within DNA can be flipped between the “right-handed” B-form and the “left-handed” Z-form. Low temperature and high salt concentration can induce this switch. An early DNA nanomachine used this mechanism to change the angle between two DNA “tiles”.
Linear motion can also be made by utilizing how DNA twists; Yang and co. created a DNA nanomacine made of a closed DNA loop attached to the arms of a Holliday junction, which is a four-arm structure containing identical DNA sequences at opposite arms. Changes in the isomerization between the matching base-pairs, by breaking at one side and then re-forming on the other, can allow this Holliday junction to migrate.
In the study by Yang and co., the intercalating dye ethidium bromide was used to change the conformation of the DNA loop. This causes the DNA strand to lengthen and partially unwind. Migration at the Holliday junction relieved the stress caused by the conformational change caused by ethidium bromide.
There is also the possibility of using these devices to report on changes in the environment; for example, conformational change can be triggered by changes in pH. In this scenario changes in pH could result in the binding of a single-stranded DNA to a DNA double helix, resulting in a triple helix. There is a possibility that, in the future, by using a combination of these sensors it might be possible to create a smart drug delivery system.
Clocked walkers
The DNA nanomachines described abvoce were based on one conformational change, but what could be achieved if multiple conformational changes were possible? A simple example consists of three single-stranded DNA anchors with a double-stranded DNA scaffold.
Each of these anchors have unique DNA sequences, which can act as “controls”, and when specific DNA fragments attach to the scaffold this can cause a conformational change to straighten a specific anchor. This can then result in a directional movement along a linear track.
Molecular motors
Molecular motors already exist, such as myosin, and these use energy generated by ATP hydrolysis as power. This has been the inspiration behind trying to create molecular motors based on DNA; for example, energy generated by breaking the phosphodiester bonds of the DNA backbone can potentially be used as a power for molecular motors.
An example of a DNA nanomachine
Ma and co. investigated the use of a DNA nanomachine which can function in a cell. This DNA nanomachine is powered by ATP. The components of the DNA nanomachine was assembled on a gold nanoparticle.
A hairpin-locked swing arm was added to the gold nanoparticle and this included a “start”. The start is triggered by an intracellular target molecule, which causes the swing arm to move. A double-stranded DNA track then responds to the moving swing arm. The authors also noted that this can be used to visualize microRNA in living cells.
Sources
Bath, J. and Tuberfield, A. J (2007) DNA Nanomachines. Nature Nanotechnology https://www.nature.com/articles/nnano.2007.104
Ma, P.-Q. and co. (2018) A highly integrated DNA nanomachine operating in living cells powered by an endogenous stimulus. Chem. Sci. https://pubs.rsc.org/en/content/articlehtml/2018/sc/c8sc00049b
Further Reading
- All Nanotechnology Content
- Nanoscience Advances in Biology
- Bionanotechnology Applications
- What are Nanoelectronic Biosensors?
- Nanoelectronic Biosensor Applications in Human Diseases
Last Updated: Dec 6, 2019

Written by
Dr. Maho Yokoyama
Dr. Maho Yokoyama is a researcher and science writer. She was awarded her Ph.D. from the University of Bath, UK, following a thesis in the field of Microbiology, where she applied functional genomics toStaphylococcus aureus . During her doctoral studies, Maho collaborated with other academics on several papers and even published some of her own work in peer-reviewed scientific journals. She also presented her work at academic conferences around the world.
Source: Read Full Article