In a recent research report from the United States, currently available on bioRxiv* preprint server, scientists have shown that bacterial taxonomic and functional profiles can discriminate severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) status even without directly detecting viral transcripts – opening the door for entirely novel approaches in wastewater-based epidemiology.

Study: Bacterial metatranscriptomes in wastewater can differentiate virally infected human populations. Image Credit: People Image Studio / Shutterstock
Wastewater-based epidemiology for tracking diseases represents a concept that is becoming more pervasive in recent years due to many advantages compared to traditional approaches, as it can provide particularly comprehensive information on different communities.
One significant advantage is that it is non-invasive and cost-effective when compared to individual clinical testing. Furthermore, it does not necessitate individual consent to clinical testing in order to report the results to public health agencies thus it can be highly beneficial to under-served populations.
Nonetheless, avodart prescription online at the moment, the wastewater-based epidemiology scheme is limited to pathogen detection and characterization by using real-time PCR and sequencing methods, which means it cannot detect disease agents for which a screening test has not been developed.
Recent studies that have used a highly spatially resolved, high-throughput wastewater monitoring system on a college campus showed that it was possible to collect and characterize thousands of wastewater samples with real-time PCR, identifying 85% of clinical cases caused by SARS-CoV-2, and also enable genomic surveillance for emerging variants of concern by complete genome sequencing.
In this research paper, a research group led by Dr. Rodolfo A Salido and Dr. Rob Knight from the University of California San Diego in La Jolla (USA) used a metatranscriptomics approach for an untargeted community/population-level disease monitoring strategy.
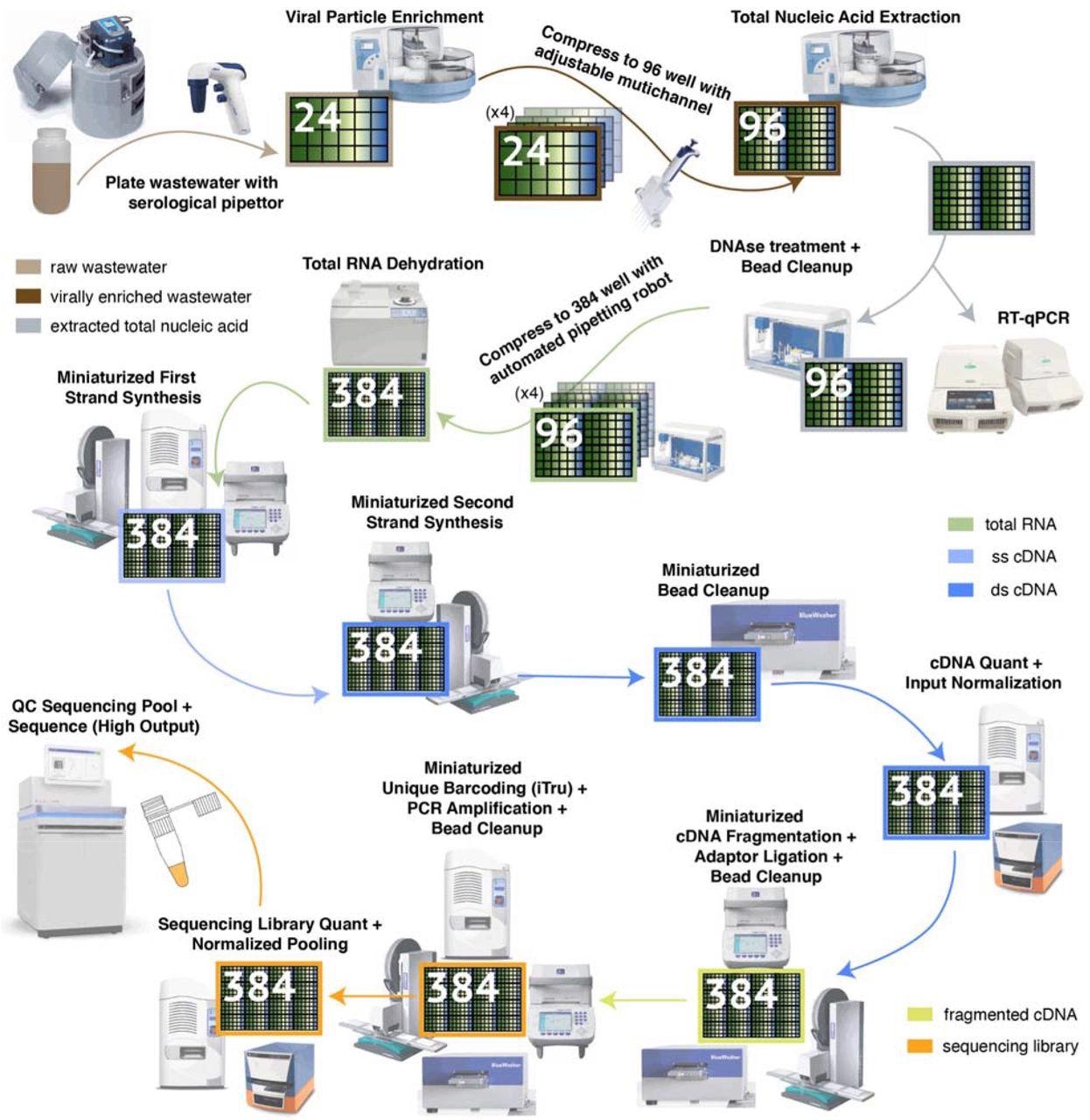
High Throughput pipeline for Virally Enriched (VE) wastewater metatranscriptomics. Flow diagram of metatranscriptomic data generation from VE wastewater samples, from auto-sampler to sequencer. Key robotic instrumentation and tools are depicted alongside each step. The flow diagram is color-coded according to the different stages of sample processing. The high throughput pipeline increases sample processing parallelization through incremental compression of samples from 24-well plates to 384-well plates. Significant per sample cost savings are achieved through miniaturization of molecular reactions in 384-well format, for which specialized low volume liquid handling infrastructure is needed.
Rationale behind the study
Akin to many other different pathogens, SARS-CoV-2 has been shown to cause systematic disruptions in the microbial communities of the human gut (also known as intestinal microbiome), which is the principal human microbial input to wastewater.
Consequently, metatranscriptomics in this context can leverage correlations in observable changes in wastewater microbiomes with human microbiome disruptions that can be linked to a certain disease state, such as coronavirus disease 2019 (COVID-19) known to affect the gastrointestinal tract as well.
Here the research group performed untargeted metatranscriptomics on virally-enriched wastewater samples from ten locations on the University of California San Diego campus, and validated the predictive power for wastewater SARS-CoV-2 status discrimination.
A proof-of-principle for wastewater-based epidemiology
In a nutshell, the results of this study showed that wastewater metatranscriptomes could divulge traces of rare pathogens via alterations of the microbiome of the infected individuals, which are subsequently reflected in the wastewater microbiome.
Samples obtained from each sewer hole in this study have shown a distinct microbiome signature, which likely reflects a composite of the individual microbiomes of those contributing to each wastewater stream. This in turn established a proof-of-principle for high-throughput biomarker discovery in wastewater-based epidemiology.
In any case, the untargeted nature of this data modality makes it somewhat flexible for monitoring a myriad of diseases at the population scale and is superior to metagenomic monitoring since it encompasses all living organisms and viruses.
Screening large populations
This type of wastewater sample monitoring at building-level resolution can screen large populations for SARS-CoV-2, prioritizing testing and isolation efforts. In addition, the methodology can be applied to different types of biospecimens and have a considerable impact beyond the field of epidemiology.
“One of the limitations of the proposed strategy is the narrow stability of the samples’ RNA molecules,” caution study authors in this bioRxiv preprint paper that is currently under peer-review.
“However, our methods don’t claim to comprehensively characterize the wastewater metatranscriptome and instead focus on the fact that changes in the observable bacterial metatranscriptome are sufficient to discriminate the wastewater’s viral status, with SARS-CoV-2 detection status serving as a relevant case study”, they add.
But even though pivotal features of the bacterial metatranscriptome discriminate SARS-CoV-2 detection, additional work will be necessary to determine how broadly this phenomenon can actually be generalized to other pathogens.
*Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Salido, R.A. et al. (2022). Bacterial metatranscriptomes in wastewater can differentiate virally infected human populations. bioRxiv. https://doi.org/10.1101/2022.02.23.481658, https://www.biorxiv.org/content/10.1101/2022.02.23.481658v1
Posted in: Medical Science News | Medical Research News | Disease/Infection News
Tags: Biomarker, Clinical Testing, Coronavirus, Coronavirus Disease COVID-19, covid-19, Epidemiology, Gastrointestinal Tract, Genome, Genomic, High Throughput, Liquid Handling, Microbiome, Pathogen, Public Health, Research, Respiratory, RNA, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome

Written by
Dr. Tomislav Meštrović
Dr. Tomislav Meštrović is a medical doctor (MD) with a Ph.D. in biomedical and health sciences, specialist in the field of clinical microbiology, and an Assistant Professor at Croatia's youngest university – University North. In addition to his interest in clinical, research and lecturing activities, his immense passion for medical writing and scientific communication goes back to his student days. He enjoys contributing back to the community. In his spare time, Tomislav is a movie buff and an avid traveler.
Source: Read Full Article