In a recent study published in the journal Advanced Science, researchers explored the dynamic changes in the respiratory tract and gut microbiota in coronavirus disease 2019 (COVID-19) patients. They observed and analyzed these changes during different disease progression stages among patients with differential disease severities.
Study: Dynamic Alterations in the Respiratory Tract Microbiota of Patients with COVID-19 and its Association with Microbiota in the Gut. Image Credit: NIAID
Background
The dynamic changes in the respiratory tract and gut microbial communities of COVID-19 patients with different disease severities could serve as a non-invasive biomarker of invasion of pathogens in the lungs or dysbiosis of the pulmonary microbiome. This data could also inform antibiotic treatment for secondary bacterial infections during COVID-19.
About the study
In the present study, researchers collected 143 sputum and 97 fecal samples from intensive care unit (ICU), non-ICU (nICU) patients, and healthy individuals. They collected sputum samples at admission, progression, and recovery stages, while fecal samples at progression and recovery stages.
The researchers calculated the Shannon index to measure the Alpha diversity within each sample, including richness and evenness measures. Then, they used the principal coordinate analysis (PCoA) dimensional reduction methods to visualize differential Beta diversity between the respiratory tract and gut samples.
Further, the researchers examined the relative abundance of genera in the microbiota of respiratory tract samples at the admission, progression, and recovery stages to analyze the dynamic changes in the respiratory tract microbiota. Multiple linear model regression analyses facilitated exploration of the differences in respiratory tract microbial composition between ICU and nICU patients at different stages of disease progression. Likewise, a relative abundance correlation analysis revealed the relationships between the microbial genera at each disease stage.
The team performed an Analysis of Variance (ANOVA) to determine the relationships between gender, age, disease severity, and clinical indices in ICU and nICU patients. Likewise, they performed a correlation analysis of the clinical indices for all the patients. They also performed a sequential redundancy analysis (RDA) for COVID-19 patients based on their blood composition and immune factors. Lastly, the team investigated genes related to COVID-19 severity using differentially expressed gene (DEG) analysis on the samples from ICU and nICU patients.
Study findings
The COVID-19 patients had a higher relative abundance of Streptococcus in the recovery stage, suggesting its association with disease progression in ICU and nICU patients. At the admission stage, the genera Streptococcus, Atopobium, Actinomyces, and Mogibacterium were significantly lower in ICU patients, whereas the genera Veillonella, Malassezia, Neisseria, and Candida were higher. In the recovery stage samples, the genera Mogibacterium, Prevotella, Atopobium, Gemella, and Bacteroides were lower in ICU patients, whereas Candida, Enterococcus, Acinetobacter, Pseudomonas, Lautropia, and Neisseria were significantly higher. Overall, Atopobium was significantly decreased in ICU patients in all stages, whereas Streptococcus and Actinomyces decreased in the admission and progression stages.
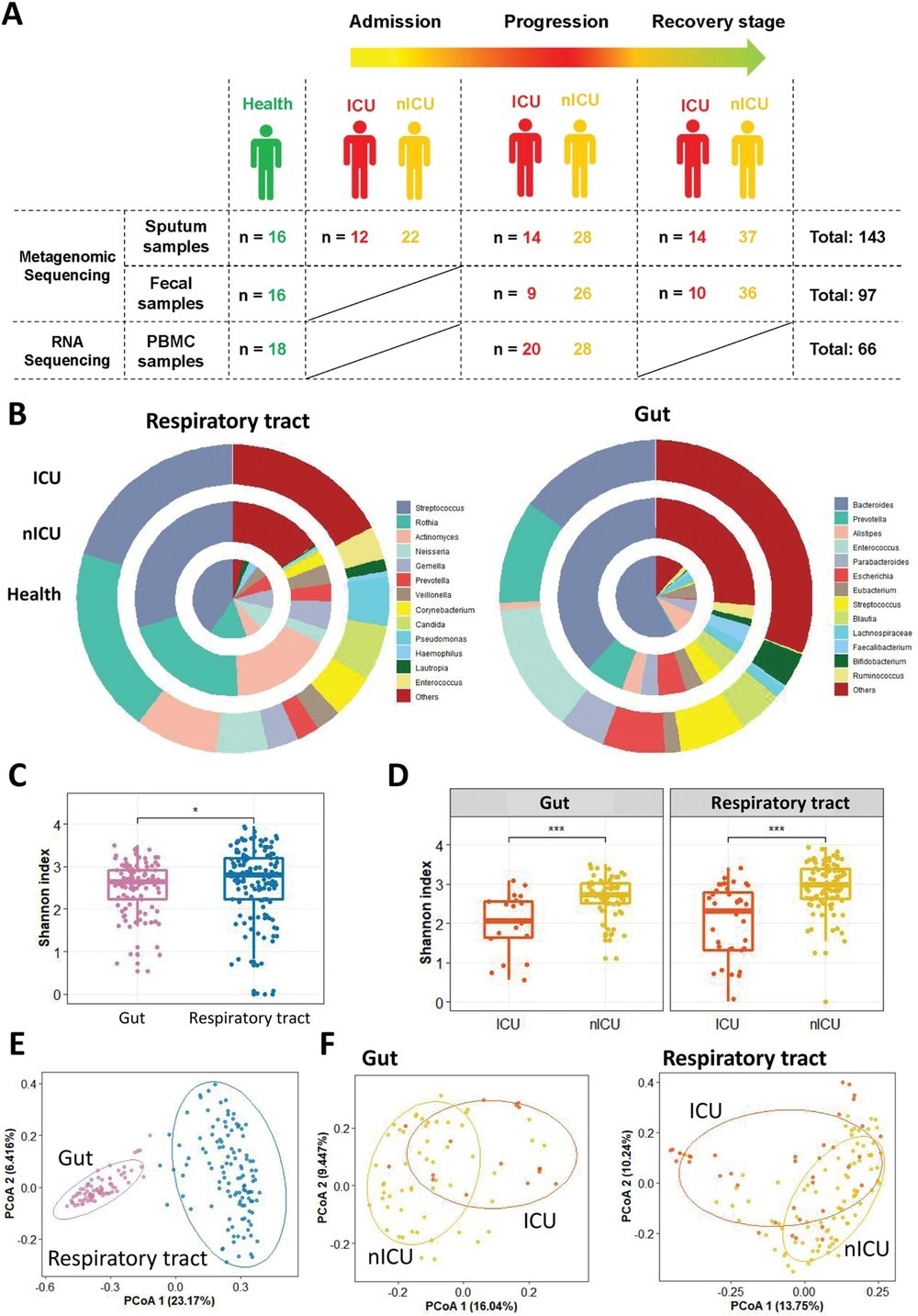
Altered respiratory tract and gut microbial compositions in patients with COVID-19. A) Overview of the experimental design. B) Microbial compositions in the respiratory tract and gut of ICU patients (n = 20), nICU patients (n = 46), and a healthy cohort. C) Comparison of alpha-diversity between respiratory tract and gut microbiota. D) Comparison of alpha-diversity between the microbiota of ICU and nICU patients in the respiratory tract and gut. * p < 0.05, ** p < 0.01, *** p < 0.001. E) First two axes of PCoA (Bray distance) for the beta-diversity of respiratory tract and gut microbiota. F) First two axes of PCoA (Bray distance) for the beta-diversity of ICU and nICU patient microbiota in the respiratory tract and gut. Group differences were tested by pairwise PERMANOVA. ICU: intensive care unit; nICU: non-ICU; PCoA: principal coordinate analysis; PERMANOVA: permutational multivariate analysis of variation; centerline, median; box limits, upper and lower quartiles; error bars, 95% CI.
The multiple linear model regression analyses confirmed that the observed differences were not due to antibiotic or corticosteroid therapy given to ICU patients during hospitalization. Instead, the reduced relative abundance of genera Streptococcus and Actinomyces was related to changes in many species, whereas only one species was responsible for the changes observed in the relative abundance of genera Atopobium, Rothia, and Veillonella.
The authors observed significantly higher levels of interleukin-6 (IL6) and IL10 cytokines in the ICU patients than in the nICU patient group, with a significant positive correlation with patients' age. Likewise, procalcitonin (PCT) levels and neutrophil counts were significantly higher in ICU patients. However, they observed a significant negative correlation between the lymphocyte count and the age of patients in both the study groups.
The blood composition RDA revealed significant correlations between the neutrophil, lymphocyte, and platelet counts and the microbial composition of COVID-19 patients. Similarly, the immune factor RDA indicated a correlation between IL10 and interferon-gamma (IFN-��) with the microbial composition of these patients.
The Spearman correlation analysis showed a negative correlation between neutrophil counts and the relative abundances of Streptococcus, Actinomyces, and Rothia in respiratory tract microbiota of COVID-19 patients. Conversely, the relative abundance of genera Neisseria positively correlated with the neutrophil count. Regarding species level correlation, the authors observed a significant positive correlation between PCT and IFN-�� levels with species Neisseria elongate, Pseudomonas aeruginosa, Streptococcus gordonii, Streptococcus milleri, and Rothia aeria, respectively.
The authors identified 914 DEGs between the ICU and nICU patient groups. In the ICU patient group, 130 and 784 genes were upregulated and downregulated, respectively. Gene ontology (GO) enrichment analysis identified 'neutrophil-mediated immunity' as one of the most enriched GO terms.
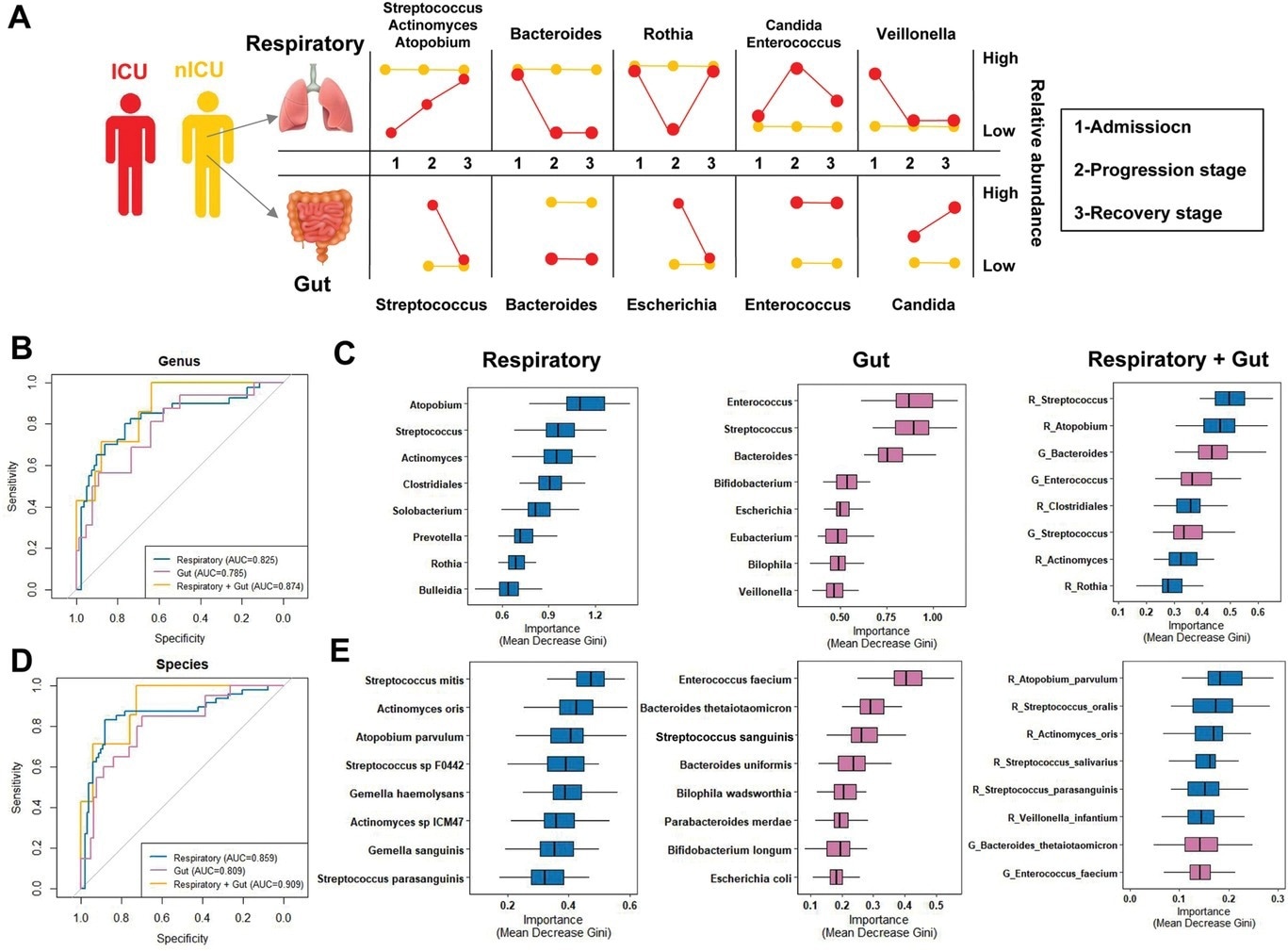
Respiratory tract and gut microbial dynamics during COVID-19 progression and their diagnostic potential for disease severity. A) Graphical representation of major microbial alterations during disease progression in the respiratory tract and gut. ROC curves showing the discriminative ability between ICU (n = 20) and nICU (n = 46) patients using the relative abundance of the respiratory tract, gut, and combined respiratory tract–gut microbiomes at the B) genus and D) species levels. Top eight important C) genera and E) species based on Gini importance according to random-forest classifiers based on the respiratory tract, gut, and combined respiratory tract–gut microbiomes. ICU: intensive care unit; nICU: non-ICU; ROC: receiver operating characteristic; centerline, median; box limits, upper and lower quartiles; error bars, 95% CI.
Conclusions
In the respiratory microbiome, Streptococcus, Rothia, and Actinomyces were the top three most abundant genera in both healthy individuals and COVID-19 patients. Bacteroides were the most abundant genus in the gut microbiota of the healthy cohort. Its relative abundance, however, decreased significantly in COVID-19 patients. Conversely, the relative abundance of Enterococcus increased considerably in the fecal samples of COVID-19 patients.
Furthermore, the authors observed a higher Shannon index for the respiratory tract microbiota. However, the ICU patients had significantly lower Shannon indices than the nICU patients for both respiratory tract and gut microbial samples, indicating an overall reduced microbial diversity in ICU patients. Furthermore, the microbial Beta diversity was significantly higher in the respiratory tract than in the gut.
Overall, the study data is invaluable as the combined respiratory tract–gut microbiota classifier could better predict COVID-19 severity. Since changes in the respiratory tract microbiota were more pronounced, these changes could serve as a prognostic biomarker for COVID-19 severity.
- Dynamic Alterations in the Respiratory Tract Microbiota of Patients with COVID-19 and its Association with Microbiota in the Gut, Yifei Shen, Fei Yu, Dan Zhang, Qianda Zou, Mengxiao Xie, Xiao Chen, Lingjun Yuan, Bin Lou, Guoliang Xie, Ruonan Wang, Xianzhi Yang, Weizhen Chen, Qi Wang, Baihuan Feng, Yun Teng, Yuejiao Dong, Li Huang, Jiaqi Bao, Dongsheng Han, Chang Liu, Wei Wu, Xia Liu, Longjiang Fan, Michael P. Timko, Shufa Zheng, Yu Chen, Adv. Sci. 2022, DOI: https://doi.org/10.1002/advs.202200956, https://onlinelibrary.wiley.com/doi/10.1002/advs.202200956
Posted in: Child Health News | Men's Health News | Medical Research News | Medical Condition News | Women's Health News | Disease/Infection News
Tags: Acinetobacter, Antibiotic, Biomarker, Blood, Candida, Coronavirus, Coronavirus Disease COVID-19, Corticosteroid, Cytokines, Diagnostic, Dysbiosis, Enterococcus, Gene, Genes, immunity, Intensive Care, Interferon, Interferon-gamma, Interleukin, Interleukin-6, Lungs, Lymphocyte, Microbiome, Platelet, Procalcitonin, Respiratory, Streptococcus gordonii

Written by
Neha Mathur
Neha is a digital marketing professional based in Gurugram, India. She has a Master’s degree from the University of Rajasthan with a specialization in Biotechnology in 2008. She has experience in pre-clinical research as part of her research project in The Department of Toxicology at the prestigious Central Drug Research Institute (CDRI), Lucknow, India. She also holds a certification in C++ programming.
Source: Read Full Article