In a recent study published on the preprint server bioRxiv*, researchers find that there is likely recombination of genome sequences between the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Delta and Omicron variants in their spike (S) protein, which can affect the vaccine landscape and their efficacy.
Study: Identification of a Novel SARS-CoV-2 Delta-Omicron Recombinant Virus in the United States. Image Credit: Kavic.O / Shutterstock.com
The importance of studying recombinant viruses
All viruses, including SARS-CoV-2, tend to undergo genomic changes over time, which may have a minor or significant impact on their infectivity, properties, and severity. Genetic changes can occur through either mutations, triphala gold side effects insertions, or recombination.
Taken together, genetic changes to viruses may lead to the accumulation of mutations, increased transmissibility, and alter the effectiveness of therapeutic vaccines. Therefore, identifying recombinant viruses may help in preparing for these events, as well as assist in the development of effective therapeutic strategies and prevention measures.
Although several studies have been published on the recombination of the SARS-CoV-2 Alpha and Delta variants, limited data is currently available on the recombination between the Delta and Omicron variants.
About the study
In the current study, nine different candidate recombinant sequences of SARS-CoV-2 were identified by Bolotie from publicly available Genbank and GISAID EpiCoVTM datasets of the mid-Atlantic part of the United States. Bolotie is an algorithm for detecting inter-clade recombinant forms, which were used to confirm recombinant genomes and the parent Delta (Clade 21J) and parent Omicron (Clade 21K) with orf1ab:2855V, 4176N, 6248S and S:95I, 142D,157-, 346K, and 501Y mutations.
To ensure that these recombinant events evolved naturally, rather than through laboratory contaminations, co-infections, or bioinformatic errors, raw read data were sequenced using Illumina and PacBio, followed by Nanopore sequencing.
Nextclade, another open-source tool, was used to identify differences in the whole genome. To this end, the nine genomes were classified as 21K (Omicron/BA.1), which were split into two fragments including the 22150 base fragment known as Clade 21J (Delta) and the other from base 22151 was Clade 21K (Omicron/BA.1.1).
The Pangolin version 3.1.20, which is a software that was developed to obtain nomenclature of SARS-Cov-2 lineages, classified the first fragment of each recombinant genome as AY.43 (Delta).
Study findings
No differentiating mutations were reported in both the parent clades between nucleotide positions 22035 and 22577. However, mutations such as Delta-associated substitutions to Omicron-associated substitutions were observed in the nine sequences, mostly between spike amino acids 158 and 339. Interestingly, the single breakpoint was different in the recombinant samples isolated from the United Kingdom.
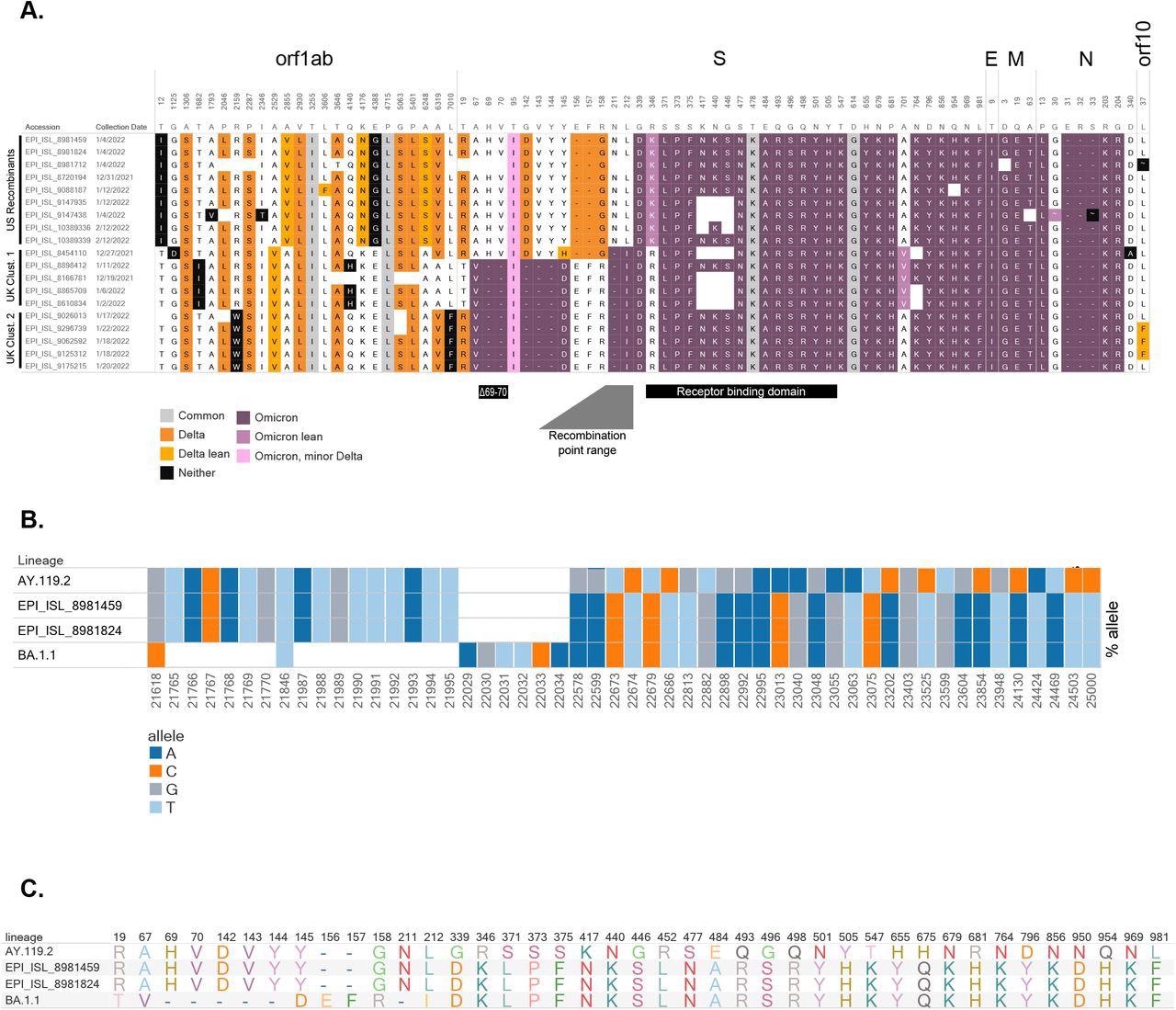
Composition of candidate recombinant virus genomes (A) Amino acid profiles of putative recombinants in the United States and the United Kingdom. Delta-associated mutations are shown in orange and omicron-associated mutations are shown in purple (see Appendix). Distinct clusters are shown, as sequences from the US appear to have recombination within S, while UK samples from two clusters show recombination before S. The BA.1.1 (Omicron) deletion associated with “S-gene target failure” (Δ69-70), the receptor-binding domain, and the range containing the recombination location is noted. (B) Bar chart illustrating the proportion of reads supporting each SNV and deletion around the recombination site for a representative AY.119.2 (Delta) genome [EPI_ISL_6811176], recombinant [EPI_ISL_8981459, EPI_ISL_8981824], and a representative BA.1.1 (Omicron) genome [EPI_ISL_9351600]. Each bar shows the proportion of reads containing the given allele (colored by A, C, T, and G) at each position for each sample. White boxes denote deletions, variants are relative to Wuhan-Hu-1. (C) Amino acid profiles within S gene for the representative AY.119.2 (Delta), the candidate recombinants, and representative BA.1.1 (Omicron) specimens.
The possibility of laboratory contamination or co-infection was ruled out. To this end, the first fragment had homology with the AY.119.2 (Delta) sequences as a result of mutations orf1ab:A2855V, orf1ab:A6248S, and orf1ab:K4176N that were common to AY.119 lineages and AY.119.2 sequences, respectively.
Over 99% frequency of Delta mutations such as C21618G, C21846T, G21987A, and deletion 22029-22034 in the 5’end was observed as compared to the AY.119.2 genome by all three sequencing methods. The coverage was over 600 times for Oxford Nanopore Technology (ONT), over 1,800 times with PacBio, and more than 1,000 times for Illumina.
The Delta origin was confirmed at the 5’ end of the S protein by the absence of BA.1.1 (Omicron) deletions at the beginning of S (21765-21770 and 21987-21995) and the characteristic Omicron “EPE” insertion at 22205 in the read data. However, the mutation profile matched with that of the Omicron representative after position 22577.
The ONT analysis revealed characteristic Delta mutations coinciding with Omicron single nucleotide variants on the same reads. Thus, the spike protein appears to be a hybrid of the SARS-CoV-2 Delta and Omicron variants, with a breakpoint at the N-terminal domain (NTD) and receptor-binding domain (RBD).
Moreover, the parent genome of recombinants was observed by splitting the candidate genomes at position 22150 and placed on the reference genome tree. Nucleotides 1-22150 clustered with Clade 21J sequences, while the other fragment of the genome clustered with 21K.
Conclusions
Overall, the current study aimed to determine recombinant genomes of SARS-CoV-2 variants and provide evidence for the possibility of contamination and/or co-infection. To this end, the researchers found that these recombinations result from substitutions common to Delta lineages at the 5’ end and Omicron lineages at the 3’ end.
The current study was the first to show recombinants between Delta (AY.119.2) and Omicron (BA.1.1) in the spike protein. Thus, the study findings may motivate others to conduct studies on laboratory and bioinformatic parameters to determine recombinant viruses.
*Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Lacek, K. A., Rambo-Martin, B. L., Batra, D., et al. (2022). Identification of a Novel SARS-CoV-2 Delta-Omicron Recombinant Virus in the United States. bioRxiv. doi:10.1101/2022.03.19.484981. https://www.biorxiv.org/content/10.1101/2022.03.19.484981v1.full.pdf.
Posted in: Microbiology | Genomics | Medical Science News | Medical Research News | Disease/Infection News
Tags: Contamination, Coronavirus, Coronavirus Disease COVID-19, Efficacy, Frequency, Genetic, Genome, Genomic, Illumina, Laboratory, Mutation, Nucleotide, Nucleotides, Omicron, Protein, Receptor, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spike Protein, Syndrome, Vaccine, Virus

Written by
Prajakta Tambe
Prajakta Tambe, Ph.D. worked at Queen’s University Belfast on a project that focused on studying ‘Role of Tregs in Acute Respiratory Distress Syndrome'. Prajakta completed a Ph.D. in August 2020 at Agharkar Research Institute, University of Pune, India. Her work aimed to develop dendrimer-based nanoparticles for the targeted delivery of MCL-1 gene-specific siRNA to bring about apoptosis in breast and prostate cancer cells and in vivo breast cancer xenograft models.
Source: Read Full Article