The coronavirus disease 2019 (COVID-19) pandemic has been caused by the rapid outbreak of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). SARS-CoV-2 has undergone mutations, which have led to the evolution of variants. Some of these variants are more virulent than the original strain and labeled as variants of concern (VOC). Due to VOCs' rapid transmission, there is a need for widespread nucleic acid testing outside of centralized clinical laboratories.
Now, a new study published on the medRxiv* preprint server describes and benchmarks SHINEv2, a Cas13-based nucleic acid diagnostic for SARS-CoV-2 detection. SHINEv2 combines temperature sample processing and lyophilized reagents to simplify the test procedure and assay distribution immensely.

Background
Frequent and widespread testing is crucial to prevent and respond to infectious disease outbreaks, such as COVID-19. Frequent diagnostic testing can aid in identifying new cases and isolating infected individuals, thereby preventing further viral spread. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) is the gold standard for COVID-19 diagnosis, but it requires specialized equipment and expertise. Lateral flow antigen-capture tests and isothermal nucleic acid diagnostics are promising alternatives for decentralized SARS-CoV-2 testing. However, they are too costly for single use and may be difficult to manufacture on a large scale. Thus, alternative diagnostic technologies that enable quick and decentralized testing are crucial to respond to current and future pandemics.
CRISPR-based diagnostics (CRISPR-Dx) are promising technologies for SARS-CoV-2 testing with minimal equipment requirements. They combine isothermal nucleic acid amplification methods and an RNA-guided CRISPR-Cas nuclease. This considerably enhances specificity and sensitivity, but at the expense of increasing assay complexity. Scientists previously developed Streamlined Highlighting of Infections to Navigate Epidemics (SHINEv1), a diagnostic assay that did not require nucleic acid extractions or custom equipment. However, SHINEv1 had certain limitations, such as frequent heating steps and requiring reagent mixtures needing cold storage. SHINEv2 is an improved version of SHINEv1 and is a fast, user-friendly, and widely deployable technology for detecting SARS-CoV-2 VOCs.
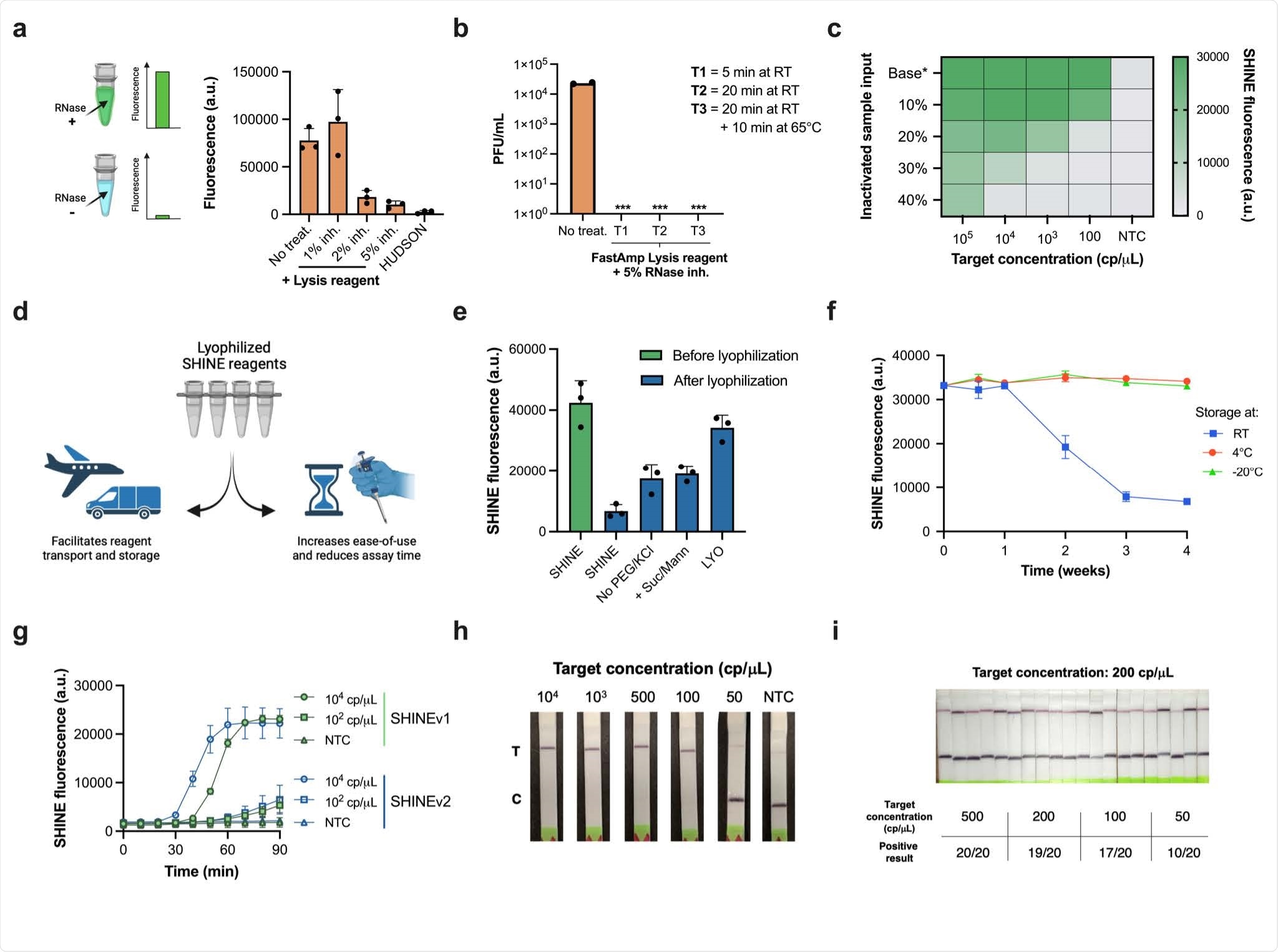
SHINEv2
As mentioned, this study develops SHINEv2, a widely deployable CRISPR-Dx for SARS-CoV-2 RNA detection. It is also capable of VOC identification from unextracted samples with a straightforward workflow, unlike previous methods. SHINEv2 also does not require a cold chain and auxiliary equipment. In addition, the assay is considerably simplified by lyophilization, which also helps in transportation and storage.
SHINEv2 is capable of being distributed overseas without a loss in performance. Further, the user-friendliness of the assay is significantly bolstered as this technology is equipment-free and uses an ambient-temperature sample lysis method. As a result, SHINEv2 involves very few steps from the user and provides a 50-fold boost in sensitivity. Another salient feature of SHINEv2 is that it perfectly aligns with RT-qPCR in samples with RNA levels above our analytical LoD of 200 copies/μL. This level of sensitivity is quite impressive as it could enable the detection of every potentially infectious individual, even those missed by antigen-capture tests.
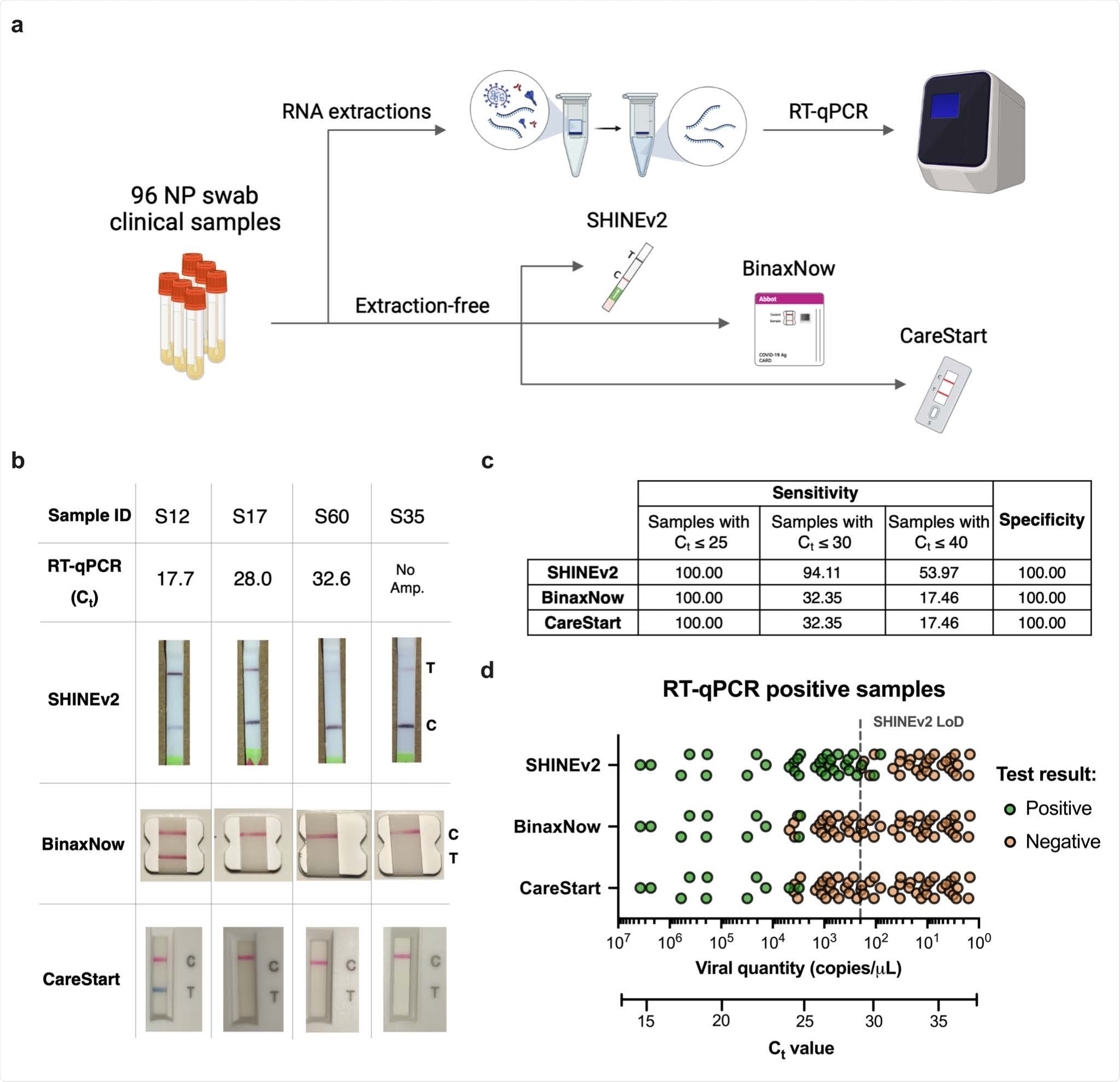
SHINEv2 is a significant improvement over previous methods as it can identify several mutations in the Alpha, Beta, Gamma, and Delta VOCs. It is also quite versatile as it can adapt to emerging viral variants and other viruses in current and future outbreaks. Therefore, SHINEv2 could inform public health responses by providing critical information.
SHINEv2 could also be used to prioritize testing and vaccine rollout in highly affected communities. It could also help doctors in selecting the right treatment for patients with severe COVID-19. Overall, scientists believe that SHINEv2 will be extremely valuable for community surveillance testing. The user-friendly and equipment-free nature of this technology makes it particularly attractive.
Conclusion
Although SHINEv2 is a significant improvement over SHINEv1, more research and advancements are needed for CRISPR-based diagnostic testing to take place in any location, including domestic settings. Ideally, such a test would not require any specialized equipment and involve a few simple, ambient temperature steps to provide a fast and accurate visual readout. Current nucleic acid diagnostics do not meet all the above-listed criteria simultaneously.
Liquid handling steps could be reduced by combining sample processing, nucleic acid amplification, and CRISPR-based detection into a single, ambient-temperature reaction. Further, the assay could be simplified, and the risk of contamination could be reduced by incorporating solution-based colorimetric readouts. Additional improvements will be needed to boost SHINEv2’s performance at ambient temperature and the addition of auxiliary proteins could bring about this change.
Collectively, these improvements could provide a critical tool in the fight against current and future pandemics. By reducing assay complexity and simplifying test distribution, researchers have taken steps towards developing a viable diagnostic tool.
*Important Notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Arizti-Sanz, J. et al. (2021) Equipment-free detection of SARS-CoV-2 and Variants of Concern using Cas13. medRxiv 2021.11.01.21265764; doi: https://doi.org/10.1101/2021.11.01.21265764, https://www.medrxiv.org/content/10.1101/2021.11.01.21265764v1
Posted in: Device / Technology News | Medical Science News | Medical Research News | Disease/Infection News
Tags: Antigen, Assay, Cas13, Cold, Cold chain, Contamination, Coronavirus, Coronavirus Disease COVID-19, CRISPR, CT, Diagnostic, Diagnostics, DNA, Evolution, Fluorescence, heat, Liquid Handling, Lyophilization, Nasopharyngeal, Nuclease, Nucleic Acid, Pandemic, Polymerase, Polymerase Chain Reaction, Public Health, Reagents, Research, Respiratory, RNA, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Transcription, Vaccine

Written by
Dr. Priyom Bose
Priyom holds a Ph.D. in Plant Biology and Biotechnology from the University of Madras, India. She is an active researcher and an experienced science writer. Priyom has also co-authored several original research articles that have been published in reputed peer-reviewed journals. She is also an avid reader and an amateur photographer.
Source: Read Full Article