Despite major progress in the global fight to end malaria, the parasite responsible for the disease has evolved resistance to every antimalarial drug deployed to date.
Experts have long worried about the emergence of drug resistance to the current frontline treatment in Africa, a region that accounts for the overwhelming majority of malaria cases and deaths. The top-line drugs, artemisinin-based combination therapies (ACTs), have been the most widely used and effective malaria treatments for 15 years. They have a fast-acting compound that rapidly clears parasites, with a longer-acting drug that kills any survivors. However, following widespread use in Southeast Asia, artemisinin resistance dramatically increased, followed shortly thereafter by resistance to nearly all ACTs in use.
Earlier this year, the first evidence of resistance to artemisinin was documented in the African country Rwanda.
A team of researchers at the Yale School of Public Health is contributing to scaling up surveillance efforts for drug-resistant malaria in Africa as the situation becomes increasingly urgent.
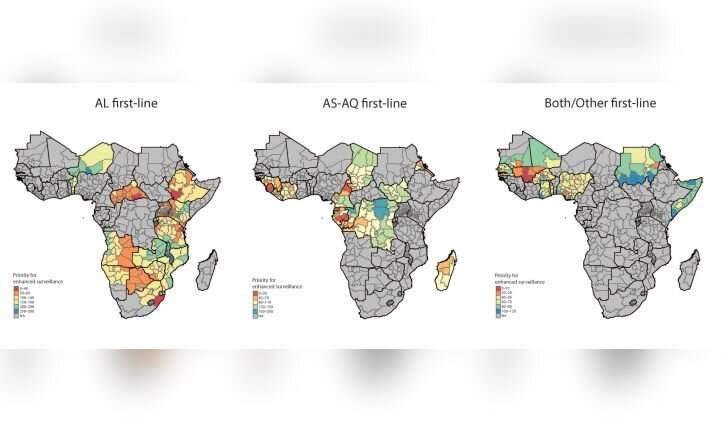
In a study published today in the journal PNAS, the team described the use of data from over 500 molecular surveys to predict the prevalence of drug resistant molecular markers across the continent, providing detailed interpolated maps for several key resistance-associated mutations across Africa’s expansive malaria-endemic region.
The study was a multidisciplinary effort at the Yale School of Public Health. It was led by Associate Professor Sunil Parikh and Associate Professor Joshua Warren. Doctoral student Hanna Ehrlich, Assistant Professor Amy Bei, and Associate Professor Dan Weinberger at the Yale School of Public Health also contributed.
The researchers found striking evidence of a continent-wide increase in molecular markers associated with reduced susceptibility to key partner drugs. Taking it a step further, they found that environmental and pharmaceutical factors, like seasonality and choice of local partner drug, influenced resistance selection over time. Finally, using advanced spatial and temporal modeling, they identified hotspots of potential or partial partner drug resistance, data which National Malaria Control Programs could use to scale up and conduct targeted surveillance.
“The maps generated from these data are striking, and show us how antimalarial drug resistance can be selected across time and space. We hope these maps will be a first step in beginning to optimize surveillance strategies for drug resistant malaria across the continent,” said Parikh.
The researchers previously characterized the state of genomic surveillance in sub-Saharan Africa over the past 15 years, utilizing data from published and unpublished sources. Their analysis found significant limitations in the coverage of genetic marker data across time and space, failing to capture the geographic scope and heterogeneity of drug resistance, as well as serious lags in the time it takes for data to be published. This work was previously published in The Lancet Microbe.
As the COVID-19 pandemic has clearly demonstrated, infectious diseases do not respect borders, and a key to containing and mitigating their impact is to begin to think critically about how we conduct surveillance, said Ehrlich.
Source: Read Full Article