Coronaviruses have been around for a long time and have caused several recent deadly outbreaks. The most recent is the ongoing pandemic of coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
A new research paper published in the Biophysical Journal describes a novel binding pocket used by a viral non-structural protein (nsp) to form an activated complex with its partner nsp. In so doing, it may have revealed a common target that may be useful in the development of a broad-spectrum antiviral drug against all coronaviruses.
.jpg)
SARS-CoV-2 nsp16
The study focuses on the working of the molecular machinery of this virus, using biophysical studies to unveil the details. The viral nsp16 is essential for viral replication because it mediates immune evasion by the virus. This protein is a 2’-O-Methyltransferase (2′-O-MTase) that forms a vital component of the viral replication-transcription complex (RTC).
In humans, the homologous protein Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase (CMTr1) carries out the same function, capping messenger ribonucleic acid (mRNA) transcripts to enhance the efficiency of protein synthesis from the mRNA template and to hide the mRNA from pathogen recognition receptors (PRRs) within the cell in which viral replication is proceeding.
When nsp16 activity is absent, viral replication is severely affected, and infectivity falls. This indicates the antiviral potential of this molecule.
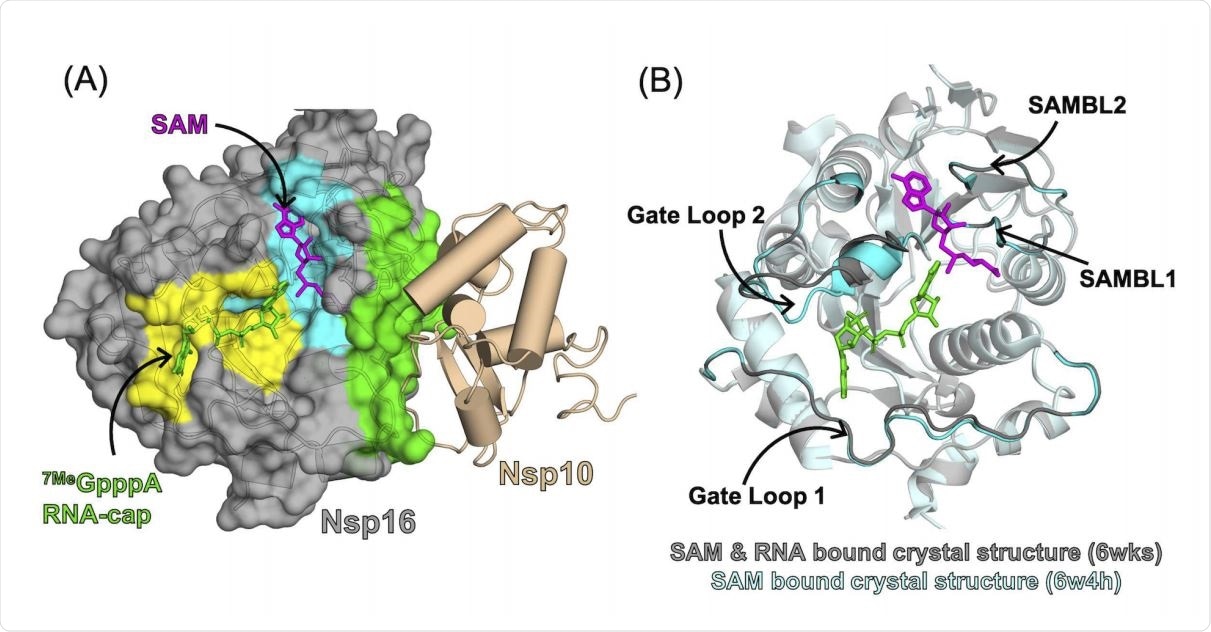
Binding partner for activation
Unlike other 2′-O-MTases in viruses or eukaryotes, nsp16 must form a binding complex with nsp10 for its activation. Failing this, it does not bind to its ligands, namely, S-adenosyl methionine (SAM) and RNA.
However, nsp10 does not show any direct binding to either of these ligands, which suggests it takes part in allosteric regulation of nsp16 to facilitate ligand binding. In other words, it stabilizes the active state of the very dynamic nsp16 structure.
This led to the hypothesis that this protein may have a cryptic allosteric pocket, which is concealed in its crystal structure. This was tested using computational modeling.
Inhibitory approaches
Inhibitors of the active site of the enzyme act by competitive inhibition of SAM binding, but since SAM acts as a cofactor for more than 200 human methyltransferases, such inhibitors will likely produce multiple off-target effects.
Another approach would be to use peptides that act at the nsp16-nsp10 interface, but such peptides are rarely shelf-stable, besides being immunogenic and requiring costly manufacturing processes.
Study aims
A third approach is envisaged in the current study, namely, using cryptic pockets. To find these, the researchers undertook a comparative study of the structure and conformational changes in the nsp16, when simulated alone or in complex with nsp10.
The simulations were then extended to earlier pathogenic coronaviruses and to the homologous human enzyme to determine if such a pocket is suitable for targeting by drug candidates.
Nsp10 enhances nsp16 pocket opening
It is known that nsp10 is necessary for the activation of nsp16. Various mechanisms have been proposed, including the ability of nsp10 to facilitate nsp16 binding to SAM and RNA by changing the conformation of the binding pocket or by directly binding with RNA. RNA binding gate loops in the complex have also been found to possess a significant opening, perhaps indicating that they are essential in the activation of nsp16, but more evidence is required.
The computational simulations of monomeric nsp16 and nsp16/nsp10 in complex were compared by a deep learning algorithm called DiffNets, which has shown itself capable of identifying the biochemical differences between different variants of the same protein by the structural changes that occur.
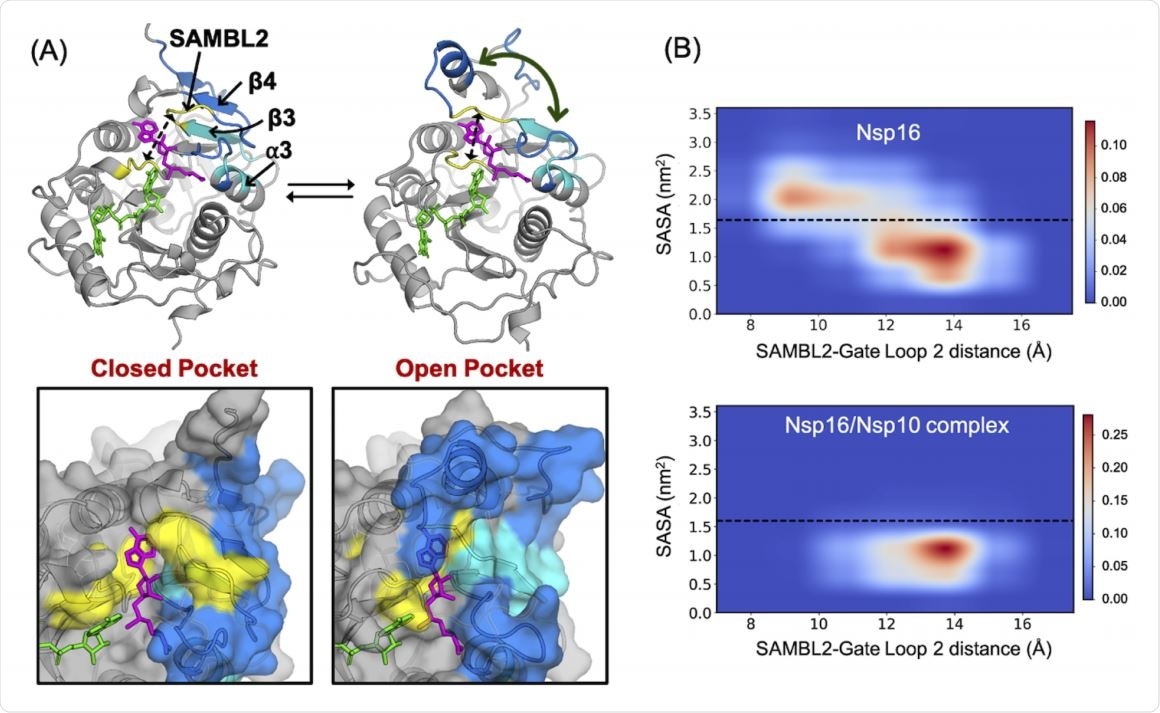
The results show that nsp10 stabilizes nsp16 conformations in order to open up both SAM- and RNA-binding pockets. This was achieved by a moving away of RNA gate loop 2 from SAM-binding loop 2, and from RNA gate loop 1, respectively.
The structural simulation that was most strongly associated with nsp16 activation closely matched a crystal structure where it was bound to both SAM and RNA.
"This is the first evidence, to our knowledge, to suggest that Nsp10 may activate Nsp16 through increasing its propensity to form a more open RNA-binding pocket."
This opening is necessary for RNA binding, of course, and suggests that both pockets tend to be open in the presence of nsp10.
They further used a statistical tool called Markov State Models (MSMs), based on molecular dynamics simulations, to represent the free energy values in the form of a network.
The MSMs showed that in the presence of nsp10, both binding pockets were stabilized in the open conformation such that they could bind SAM and RNA. This was through a marked increase in stiffness of the chief binding components, especially the SAM binding loops and RNA gate loops.
This results in gate loop 2, which contacts both SAM and RNA, being out of direct contact with nsp10. Though surprising, this points to robust allosteric effects.
Thus, nsp16 is more likely to be in open states capable of binding both ligands when nsp10 is present.
In other words, "Nsp10 aids SAM and RNA binding by preventing the collapse of SAM and RNA binding gate loops."
Nsp16 cryptic pocket could provide potential drug target
The current analysis also provides multiple structural simulations of the soluble protein at different temperatures, allowing cryptic pockets – closed and invisible – in the crystal structure to be identified. These can present unique binding sites rather than those conserved among functional homologs, thus allowing specific targeting of the interaction without off-target effects.
Using the Exposons algorithm that picks up residues that show cooperative changes in solvent exposure, they found a concave cryptic pocket formed by β3 and β4 strands, comprising 11 residues altogether. This pocket can be opened to stabilize the inactive state of the protein complex.
Several residues in this pocket make direct contact with nsp10 in the nsp16/nsp10 complex and will be displaced when the β3-β4 pocket opens. This results in the inhibition of complex formation.
This concave pocket is also easier to target with small molecule inhibitors.
"We propose that targeting the β3-β4 pocket with a small molecule could inhibit Nsp16's activity by preventing SAM binding or preventing association with Nsp10."
Conserved cryptic pocket among coronavirus nsp16
This pocket was explored for its conservation in other coronaviruses. First, they found that this pocket sequence is completely conserved in SARS-CoV and with only two mutations in MERS-CoV.
However, the sequence is different from that of the human CMTr1. Thus, even if this cryptic pocket is formed by the latter, selective inhibition is likely to be possible, though the former is unlikely.
"Taken together, features of the β3-β4 cryptic pocket in coronavirus homologs of Nsp16 appear sufficiently similar to each other and dissimilar to CMTr1 to make for a promising target for pan-coronavirus inhibitors."
What are the implications?
This modeling study's findings unravel some of the mechanisms by which nsp16 is activated and suggests a potential inhibitory target for this key component of viral replication, susceptible to small molecules. The study also shows that while this cryptic pocket is conserved among other pathogenic coronaviruses, but not the human homolog, CMTr1.
The cryptic pocket thus presents "potential for development of a pan-coronavirus, broad-spectrum inhibitor that may be efficacious against COVID19 and yet unseen coronavirus outbreaks."
- Vithani, N. et al. (2021). SARS-CoV2 Nsp16 activation mechanism and a cryptic pocket with pan-coronavirus antiviral potential. Biophysical Journal. https://doi.org/10.1016/j.bpj.2021.03.024. https://www.cell.com/biophysj/fulltext/S0006-3495(21)00254-X
Posted in: Medical Research News | Disease/Infection News
Tags: AIDS, Antiviral Drug, Cell, Coronavirus, Coronavirus Disease COVID-19, Deep Learning, Enzyme, Eukaryotes, Homologous, Ligand, Manufacturing, MERS-CoV, Methionine, Molecule, Nucleoside, Pandemic, Pathogen, Peptides, Protein, Protein Synthesis, Research, Respiratory, Ribonucleic Acid, RNA, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Structural Protein, Syndrome, Transcription, Virus

Written by
Dr. Liji Thomas
Dr. Liji Thomas is an OB-GYN, who graduated from the Government Medical College, University of Calicut, Kerala, in 2001. Liji practiced as a full-time consultant in obstetrics/gynecology in a private hospital for a few years following her graduation. She has counseled hundreds of patients facing issues from pregnancy-related problems and infertility, and has been in charge of over 2,000 deliveries, striving always to achieve a normal delivery rather than operative.
Source: Read Full Article